Databases and resources for rice genomics
We have developed databases, bioinformatics tools and genomics resources to complement our efforts in structural and functional characterization of the rice genome, to provide fundamental information for understanding the biology of rice and other cereal crops, and to facilitate the development of novel strategies for crop improvement. Central to these genomics efforts is the Rice Annotation Project Database (RAP-DB) providing comprehensive information on the rice genome including functional annotation for the gene models. A wide range of resources encompassing genome sequence data, genetic maps, molecular markers, insertional mutations, gene expression profiles, proteome, and the integration of these genomics data have been developed providing various types of information that can be used for complete understanding of rice biology.
Rice has a large base of genetic resources and knowledge of the sequence of specific genes will pave the way in maximizing 42,000 accessions representing natural genetic variation and germplasm of the rice species in the NIAS Genebank. We also provide access to genomics resources in the form of cDNA clones, DNA markers, cloned genomic DNAs in bacterial artificial chromosomes (BAC) and P1-artificial chromosomes (PAC).
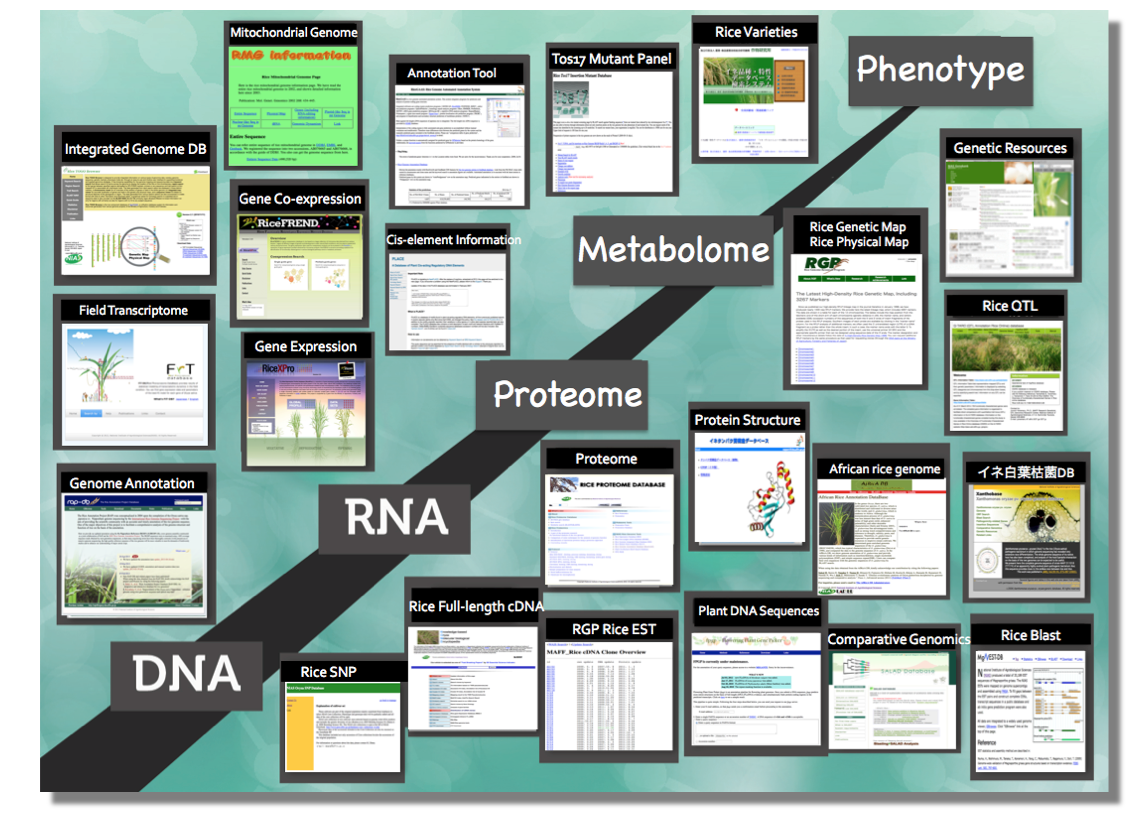
Rice genomics databases
- RAP-DB: Rice Annotation Project Database
- Rice TOGO Browser: Integrated rice genomics browser
- RiceXPro: Rice Gene Expression Profiling Database
- RiceFREND: Rice Functionally Related Gene Network Database
- FiT-DB: Field Transcriptome Database
- KOME: Rice Full-length cDNA Database
- Tos17: Rice Tos17 Insertion Mutant Panel Database
- Q-TARO: QTL Annotation Rice Online Database
- RiceGAAS: Rice Genome Automated Annotation System
- RiceTOGO Browser: Integrated rice genomics database
- AfRicA DB: African Rice Annotation Data Base
- RMG Information: Rice Mitochondrial Genome Database
- Rice Proteome DB: Rice Proteome Database
- RMOS: Rice Microarray Opening Site
- SALAD: Genome-wide database of similarity clustering based on distribution patterns of motifs
- Barley Gene Expression DB: Full-length cDNA sequences from malting barley
- DAIZUbase: Soybean genome database
- PLACE: A Database of Plant Cis-acting Regulatory DNA Elements
- Flowering Plant Gene Picker: Annotation pipeline for flowering plant genomes
- Database of Rice Cultivars and Traits (in Japanese only)
- MgNEST-DB: Magnaporthe grisea EST database
- Xanthobase: Xanthomonas oryzae pv. oryzae genome database
