 |
||
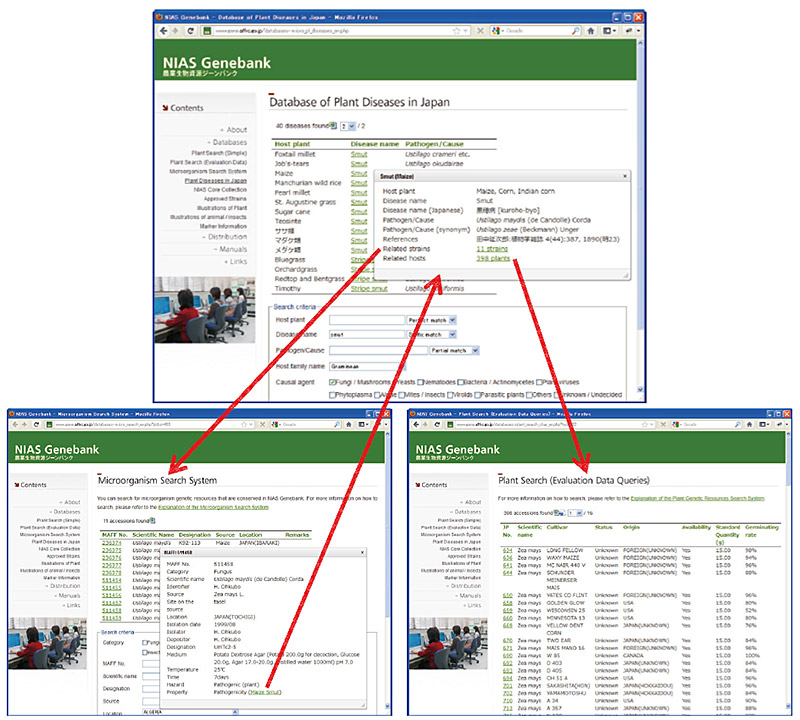
| Fig. 1. Database search interface The database is provided with a search interface that allows a user to specity the search conditions for a particular plant disease and execute a search (upper panel). The search results include links to other pathogens related to the disease (lower left) and other plant hosts of the pathogen (lower right). |