

We have currently emphasized the
characterization of microbial components of agro-ecosystems and
the methodology for the effective management of the ecosystem.
Taxonomy and systematics: Several plant pathogenic fungi and
insect parasitic nematodes were isolated and identified as new
species. A new reference system for identifying pathogenic
bacteria was developed and released for use. Rice grassy stunt
virus, a suspected member of the Tenuivirus group, was
found to have a weak similarity in the genome structure with rice
stripe virus, the type virus of the group. Insect parasitic
nematodes were evaluated for their potential value as biological
control agents.
Pathology and ecology of pathogens: a PCR method was developed for detecting some species of plant pathogens. The gene related to toxin production was isolated from some species of Pseudomonas and was confirmed in relation to their pathogenicity or pathotypes. The population structures involved in the morphological, physiological and genetic diversity in microbial flora in acidified field and oxidized soils were analyzed in relation to the complicity in beneficial and harmful organisms.
Molecular biology of microorganisms: The toxin-resistant gene (argK) from P. syringae pv. actinidiae was introduced into kiwi plants,and expression of the gene was confirmed. Several chitinase genes of Streptomyces lividuens were cloned and mapped on the chromosome. For the use of microorganisms in the remediation of environments contaminated with chlorinated aromatics, the structure and expression of the degradative genes of Pseudomonas sp. was analyzed.
Topic1
Specific Detection of Burkholderia plantarii and B. glumae by PCR Using Primers Selected from 16S-23S rDNA Spacer Regions
B. plantarii and B. glumae cause bacterial seedling blight in rice and bacterial seedling rot in rice, respectively, which are serious diseases in the nursery boxes for the paddy fields in Japan.
Polymerase chain reaction (PCR)has been developed for detecting various plant pathogenic bacteria, because it is a more rapid, sensitive,and specific method than conventional ones. The objective of this study was to design and apply PCR primers targeting the spacer region between 16S and 23S rRNA genes of B. plantarii and B. glumae for specific amplification and detection of these pathogens in field samples.
The spacer region between 16S and 23S rRNA genes of Burkholderia spp. were sequenced. On the basis of the sequence data, two pairs of PCR primers, PL-12f and PL-11r for B. plantarii and GL-13f and GL-14r for B. glumae, were designed for specific amplification. An approximately 180-bp fragment was amplified from all the 45 isolates of B. plantarii derived from different origins, by using PL-12f and PL-11r as primers, and no PCR products were obtained from other bacteria (Fig.1).
Primers GL-13f and GL-14r amplified approximately 400-bp fragments of all the 20 isolates of B. glumae from different origins,and no PCR products were obtained from other bacteria. The PCR procedure for detecting these two pathogens can be performed in the same condition. Detection sensitivity of PCR with each pair of our primers was 10 to 100 CFU per PCR reaction. PCR products were also obtained using the same primers in PCR from rice seedling samples infected with B. plantarii or B. glumae (Table 1). The pathogens in rice samples can be detected for 5 or 6 hours by the PCR method.
Topic2
A Chlorocatechol Degradative Gene Cluster on a Plasmid of Alcaligenes eutrophus NH9 and Insertion Sequence-like Elements Involved in the Rearrangement of the Gene Cluster
Pollution by chlorinated aromatic compounds presents one of the most serious environmental problems worldwide today. However, some of these compounds are degraded by microorganisms in soil and water. In the oxidative bacterial degradation of chlorinated aromatics, these compounds are transformed to chlorocatechols, which are then degraded through modified ortho-cleavage pathway. We elucidated the structure of a chlorocatechol degradative gene cluster on a plasmid of Alcaligenes eutrophus ( Ralstonia eutropha )NH9 and found a pair of highly homologous insertion sequence (IS)-like elements which were involved in the rearrangement of the degradative gene cluster.
A.eutrophus NH9 which degrades 3-chlorobenzoate (3-CB) was isolated from soil in Japan. The genes responsible for 3-CB degradation, carried on a plasmid (pENH91), were cloned. Sequence analysis revealed an operon structure homologous with those of the modified ortho-cleavage pathway. Among them the genes of NH9 (named cbnR-ABCD)showed strikingly high homology with tcbR-CDEF genes on plasmid pP51 of the 1,2,4-trichlorobenzene degrading bacterium Pseudomonas sp. P51 isolated in the Netherlands (95.6%-100% identity in amino acids level). The difference of the utilizable chloroaromatics as growth substrates between the two strains seemed to be due to the difference in available upper pathway enzymes that convert the substrates into chlorocatechols. This also illustrates an economical method of bacteria to acquire an ability to degrade a new xenobiotic compound by recruiting an available set of genes.
Two highly homologous sequences of 2.5kb were found to be present as a direct repeat at both sides of the degradative gene cluster (Fig.2, DR1, DR2) by the spontaneous loss of the cluster from the plasmid by homologous recombination. During repeated subculturing of NH9 on liquid media with 3-CB, the culture was taken over by a derivative strain, in which the degradative plasmid carried a duplicate copy of the 12.5-kb region that contained the degradative genes. Duplication of the degradative genes also seemed to have been mediated by reciprocal recombination between the direct repeat sequences. Nucleotide sequence analysis of one of the homologous fragments (DR2) revealed a structure of IS,and relatedness to IS1326 which belonged to IS21 family.
The high homology between the two clusters, cbnR-ABCD of strain NH9 and tcbR-CDEF of strain P51, suggested horizontal transfer of the clusters as a unit. This hypothesis is supported by the composite transposon-like structure consisting of cbnR-ABCD genes and IS-like elements on pENH91. The distant locations where the two strains had been isolated suggested possible world-wide distribution of this cbn (tcb) type of gene clusters. The ability of cbn gene clusters to degrade a broad range of chlorocatechols including 3,4,6-trichlorocatechol will be useful in application with genetic engineering.
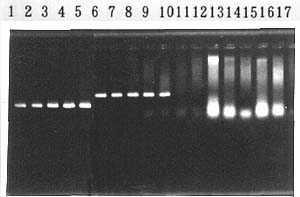
Fig.1 PCR product amplified from Burkholderia glumae (299bp) plantarii (322bp)
1-5: B. glumae, 6-10: B.plantarii, 11-12: B.gladioli, 13: P. cepacia, 14: B. caryophylli, 15: B. andropogonis, 16: B. solanacearum, 17: B. corrugata
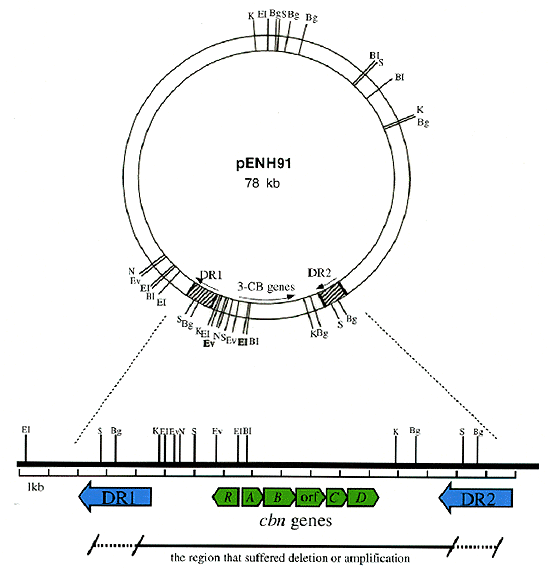
Fig.2 Restriction map of pENH91 and the region of the degradative genes.
Restriction sites are abbreviated as follows: BI, BamHI; Bg, BglII; EI, EcoRI; Ev, EcoRV; KpnI; N, Nhel; S, Sacl.