

Our main research is on the behavior of pesticides in the environment and on the pesticide resistance in phytopathogenic fungi and insect pests. On evaluation of methyl bromide emission to the atmosphere in soil fumigation, field studies showed 30-45% loss of methyl bromide used in Japan. Volatilization rates of pesticides from flooded water in agricultur al fields ranged from 3-26% of applied amount in the test period of 9 days. In surveying pesticide residues in the surface water of rivers in farm areas,herbicides for paddy field were detected at ppb levels during May through early July. To clarify the behavior of herbicide-degrading microorganisms in paddy field, we developed a new sensitive detection method using HPLC-MS/MS (HPLC: high performance liquid chromatography, MS: mass spectrometry), and a new material (woody charcoal)suitable for trapping and accumulating them. In the research on pesticide resistance, induction of systemic acquired resistance to scab was found in cucumber plants treated with a newly developed non-fungitoxic compound. Antifungal compounds were extracted from the treated plants and their chemical properties are currently under investigation. The monitoring of insecticide resistance of the common cutworm, Spodoptera litura, was established. The male adults captured in a sex-pheromone trap were used in this method.
Topic1
Predicting Pesticide Behavior in a Lowland Environment Using Computer Simulation
Studies were conducted to be able to come up with a model for predicting pesticide behavior in the paddy field. The prediction of herbicide residues in surface soil (0-1cm) and paddy water in the field by computer simulation appeared to be closely related with both the herbicidal activity and environmental safety.
As shown in Fig.1, the model has two compartments with the first compartment further subdivided into two sub compartments, namely paddy water and upper soil layer (0-5mm). The second compartment is the lower soil layer (5-10 mm). The Calculation of the simulation model requires 15 parameters. The model was evaluated by comparing the actual data obtained from the field experiment with simulated data based on 2 sets of parameter. One set of parameters was obtained from the literature while the other was obtained from laboratory measurements.
Pretilachlor concentration in paddy water in the field was quite accurately predicted until around 6 weeks by the model. There was no considerable difference in simulation results between literature parameters and measured parameter (Fig.2). Pretilachlor residues in surface soil (0-1 cm) in the paddy field was accurately predicted up to 7 weeks after herbicide application by the model using measured parameters. On the other hand,the model using literature parameters underestimated pretilachlor residues beyond 3 weeks after application (Fig.3).
Pesticide behavior in paddy field would be more accurately predicted by computer simulations, if "sensitive parameters", e.g.,degradation rates of pesticide in soil and adsorption coefficients (K), are obtained by laboratory experiment which are based on actual paddy field conditions.
Topic2
DNA-based approaches for diagnosis of fungicide resistance in phytopathogenic fungi
The long time required for testing fungicide resistance by traditional methods interferes with quick and accurate decision making to minimize the risk of control failure with the fungicide. DNA-based technology can be used to detect fungicide resistance once the mechanism of resistance has been determined at a molecular level.
The resistance mechanism of Venturia nashicola, the scab fungus of Japanese pear, against benzimida-zole fungicides has been well characterized. In this fungus,four phenotypic responses to the fungicide carbendazim were identified as follows: sensitive, weak resistance, intermediate resistance and high resistance, and these phenotypes were each governed by an allelic series in a single Mendelian gene.
Sequence analysis of the PCR-amplified β-tubulin gene in V. nashicola suggested that one base change at either codon 198 or codon 200 led to resistance in field strains. Substitution of glutamic acid (base sequence: GAG) at codon 198 in β-tubulin by either alanine (GCG) or lysine (AAG) resulted in high resistance to carbendazim. The former mutation was involved in negatively correlated cross-resistance to the N-phenylcarbamate fungicide diethofencarb, however, the latter caused double resistance to diethofencarb. In a strain showing intermediate resistance to carbendazim, the amino acid at codon 200 was altered from phenylalanine (TTC) to tyrosine (TAC).
Allele-specific PCR (ASPCR) primers were designed based on the sequence difference and used for the diagnosis of resistance. When a purified genomic DNA or PCR-amplified β-tubulin gene fragment was used as a template, the mutation in codon 198 or codon 200 was each identified by this method (Fig.4). Single-strand conformation polymorphism (SSCP) of DNA was further applied for the identification of the resistance. PCR products of the β-tubulin gene were analyzed using capillary electrophoresis. The mutation in codon 198 or codon 200 was identified as a difference in migration time of the DNA sample on a poly-acrylamide gel (Fig.5).
It is necessary to reduce fungicide applications so that resistance problems can be minimized. Increasing use of modern diagnostic tools to detect and quantify diseases and to identify fungicide resistance will play an important part in reducing the input of chemical crop protectants to the environment.
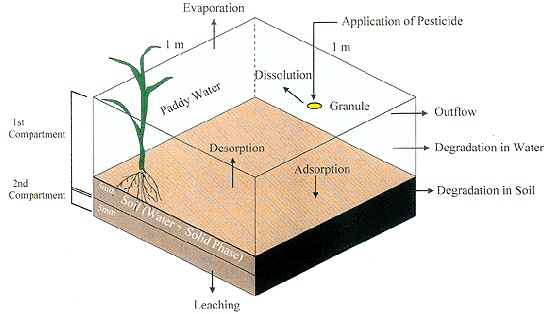
Fig.1 Compartmental Model for Pesticide Behavior in Paddy Field
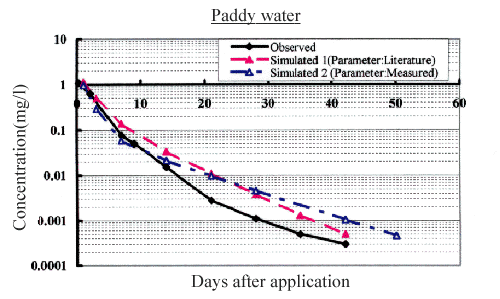
Fig.2 Observed vs simulated value of pretilachlor concentration in paddy water at NIAES field station
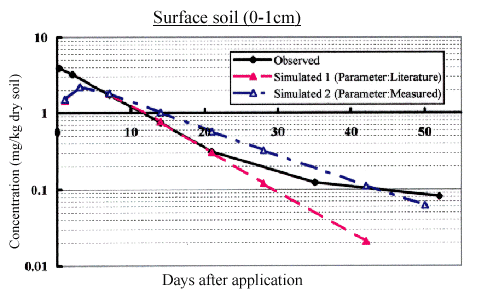
Fig.3 Observed vs simulated value of pretilachlor consentration in surface soil (0-1 cm depth) at NIAES field station
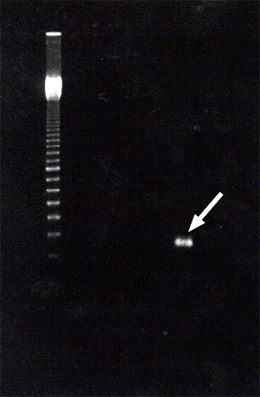
Fig.4 Electrophoresis pattern of β-tubulin gene amplified by ASPCR Arrow shows a specific band of the DNA fragment obtained from a highly carbendazim-resistant strain.
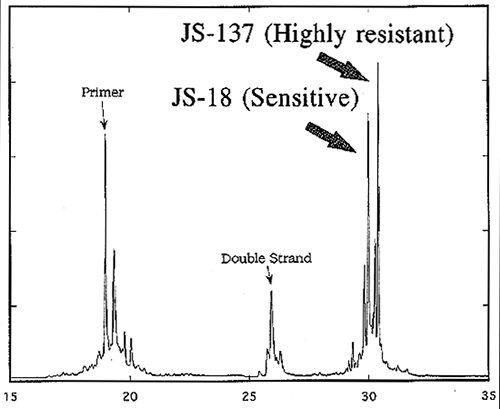
Fig.5 Detection of one base change in β-tubulin gene by SSCP analysis.
Mixture of DNA samples from both highly carbendazim-resistant and -sensitive strains was analysed.