Mapping the genes that comprose the brown planthopper genome
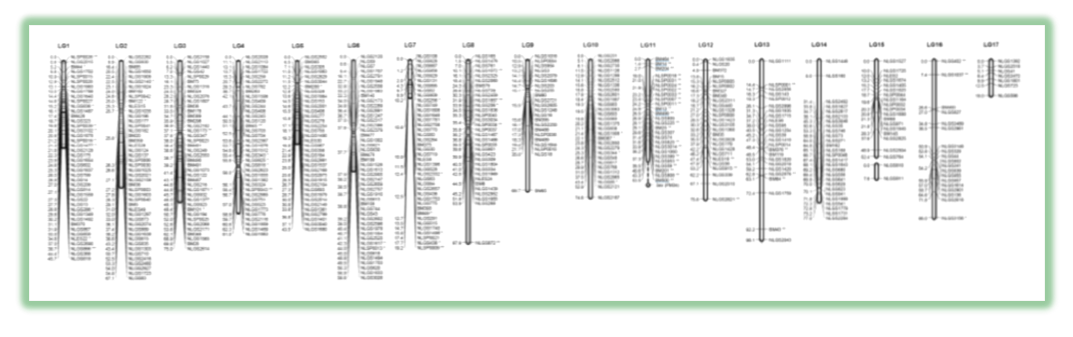
The first genetic linkage map for brown planthopper (BPH, Nilaparvata lugens), a major insect pest of rice, was developed. The linkage map was constructed by integrating linkage data from two backcross populations derived from three inbred BPH strains. The consensus map consists of 474 simple sequence repeats, 43 single-nucleotide polymorphisms, and 1 sequence-tagged site, for a total of 518 markers at 472 unique positions in 17 linkage groups.
Inbred lines were established for three BPH strains with different genetic backgrounds (Hadano-66, Chikugo-89, Koshi-10). Two backcrossed populations were generated and used as the basal materials for developing molecular markers and genetic linkage maps. SSR (simple sequence repeat) markers and SNP (single nucleotide polymorphism) markers were designed by analyzing genomic sequences and EST (expressed sequence tags) of BPH. SSR and SNP markers that are polymorphic between inbred lines were selected and their individual genotypes in backcrossed populations were detected. Recombination values between markers were calculated to determine linkage groups and generate a genetic linkage map.
The genetic linkage map and molecular markers for BPH are essential resources for genetic analyses of genes controlling agriculturally important traits of BPH, such as insecticide resistance and virulence to the resistant rice varieties.
- Overview of BPH genome research
- BPH Databases and Resources
- BPH genome and research on insect resistance genes
