
| |
Fig. 1. Modification of OASA2 gene via GT | |
 | 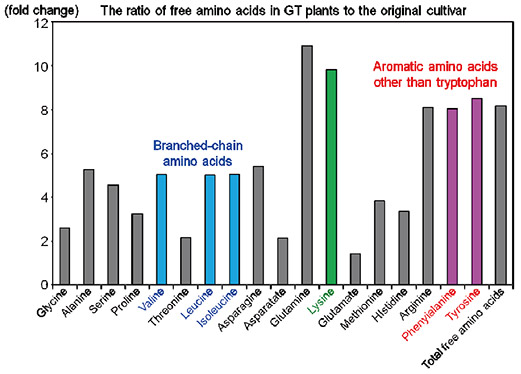 |
| Fig. 2. Free tryptophan contents in leaves and seeds of GT plants The free tryptophan content in leaves and mature seeds of GT plants is higher than in the original cultivar. | Fig. 3. The ratio of free amino acids in seeds of GT plants to that in the original cultivar The contents of free amino acids other than tryptophan in mature seeds of GT plants are higher compared to the original cultivar. Overall, the total amount of free amino acids is >8-fold higher than in the original cultivar. |