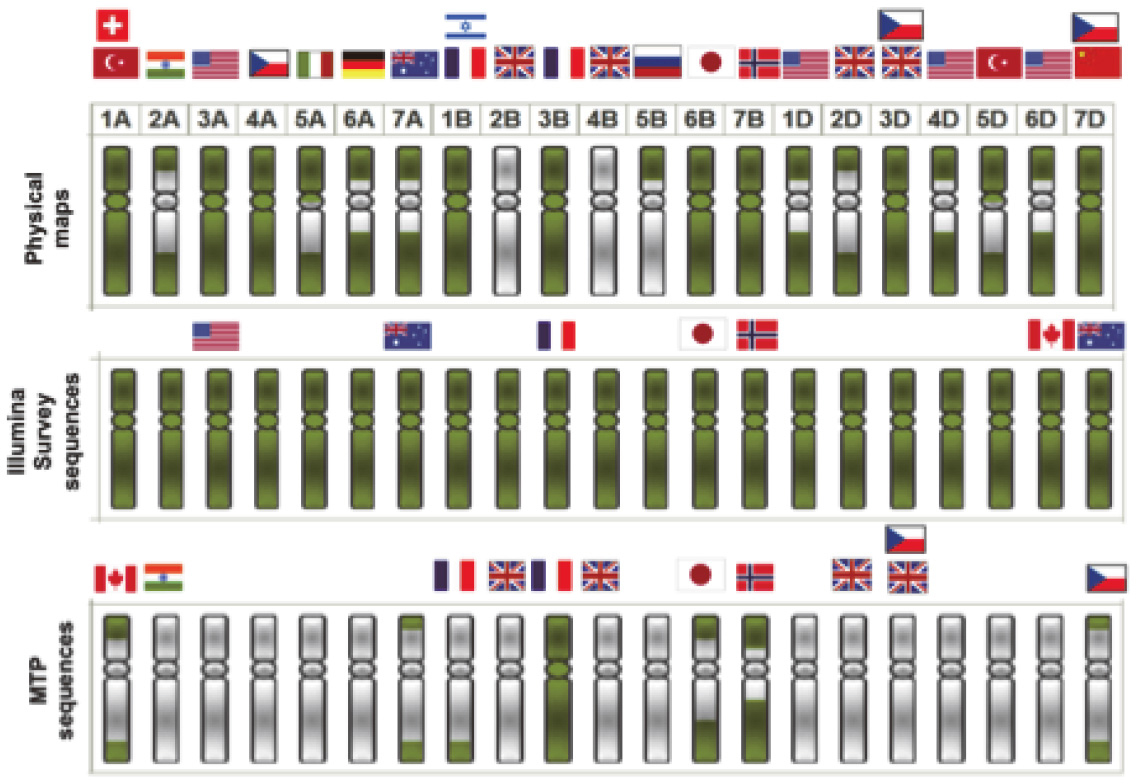
 |
| Fig. 1. Current status of genome sequencing in terms of physical map construction (top panel), draft genome sequencing (middle) and reference genome sequencing (bottom). The state of progress is shown in green |
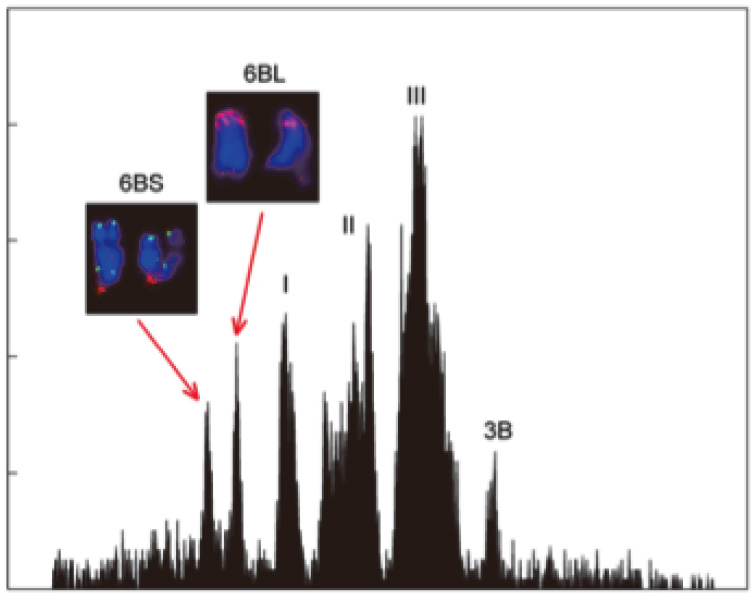 |
| Fig. 2. Isolation of wheat chromosome 6B by flow cytometry. |
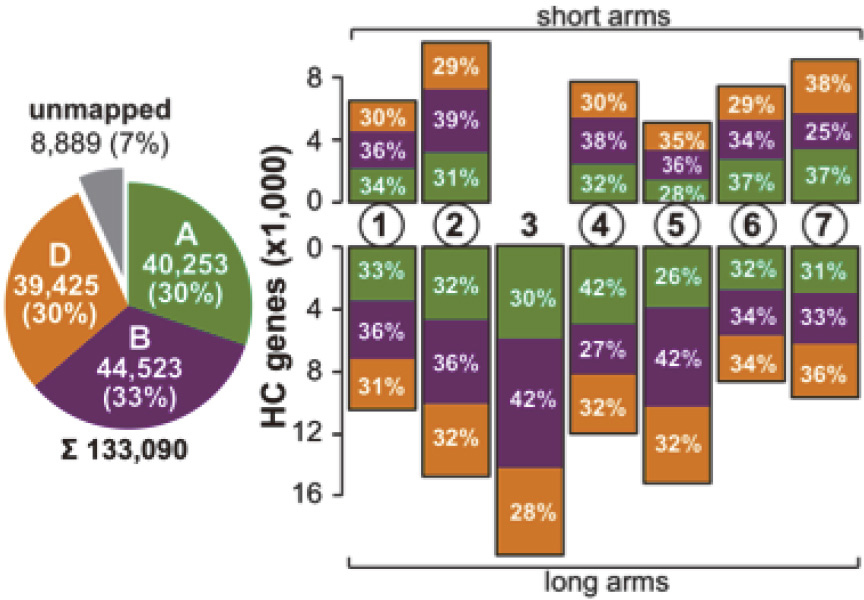 |
| Fig. 3. Distribution and total number of HC (high confidence) bread wheat genes identified on the A (green), B (purple), and D (orange) subgenomes. |
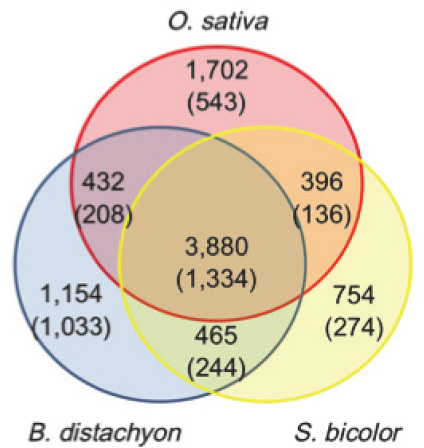 |
| Fig. 4. Distribution of chromosome 6B genes with significant similarity to O. sativa, B. distachyon and S. bicolor. |
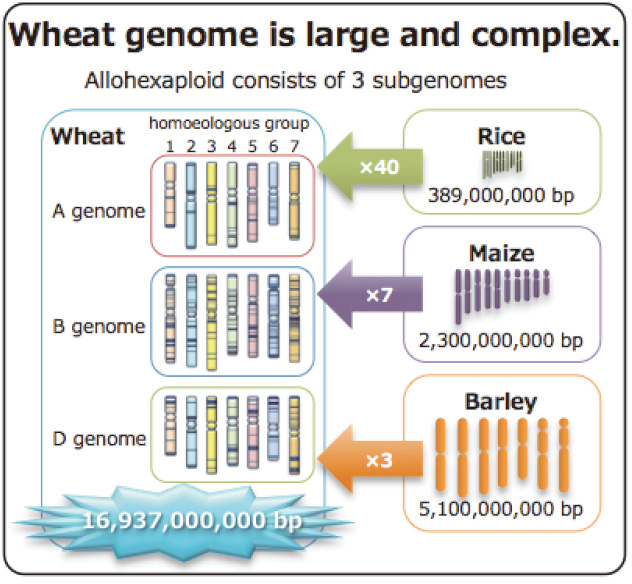 |
| Fig. 5. Comparison of wheat and other cereal crop genomes. |