 |
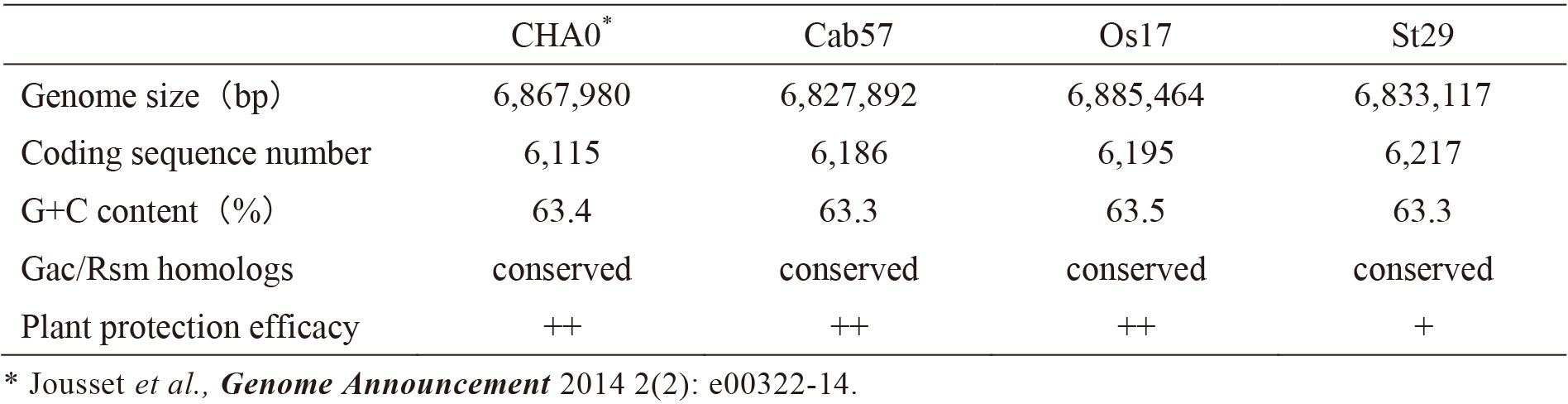
| Table 1. General genomic features and comparison of plant protection efficacy of Pseudomonas protegens CHA0, P. protegens Cab57, Pseudomonas sp. Os17, and Pseudomonas sp. St29. |
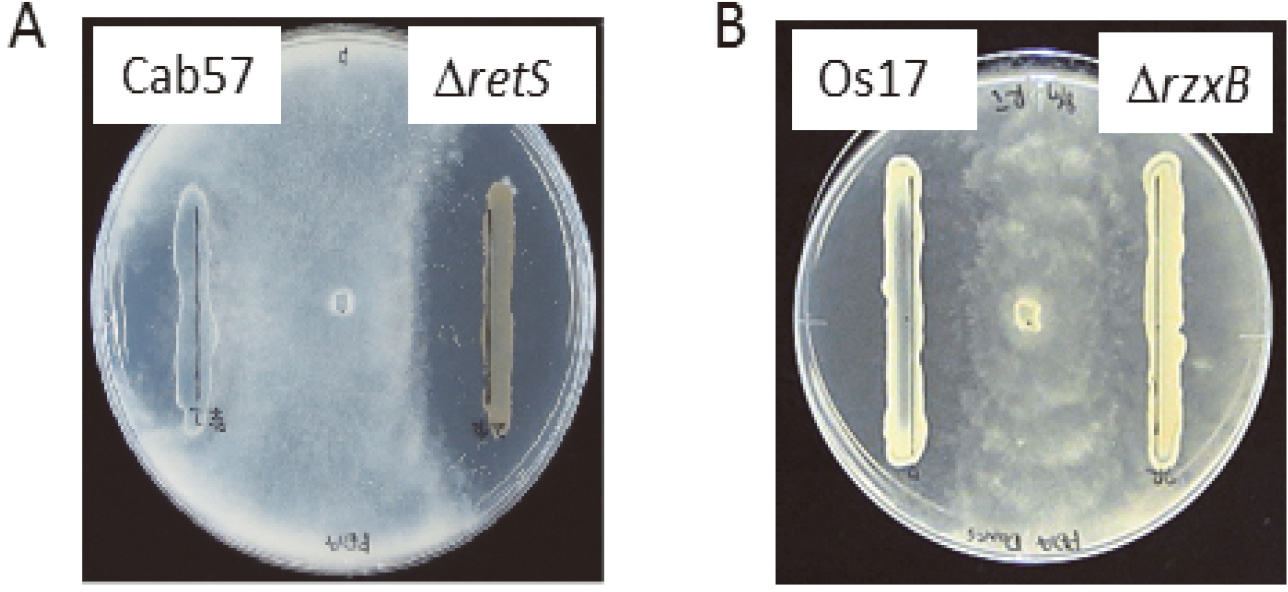 |
| Fig. 1. Antibiotic activities of Pseudomonas strains toward Pythium ultimum. (A) Antibiotic activity of P. protegens Cab57 wild type was enhanced by knocking-out the retS gene. (B) Antibiotic activity of Pseudomonas sp. Os17 wild type was attenuated by knocking-out the rzxB gene. |